|
||||||||||||||||||||||||||||||
| [发表评论] [本类其他产品] [本类其他供应商] [收藏] | ||||||||||||||||||||||||||||||
| 销售商: 蓝景科信河北生物科技有限公司 | 查看该公司所有产品 >> |
技术特点:
- 捕获效率高,覆盖范围广,对被子植物的353个单拷贝核基因进行定向捕获测序
- 适用多种样本类型,节约时间成本和经费成本
应用领域:
- 系统发育学
- 群体遗传学
- 古遗传学
- 侧翼区捕获
- 内含子测序
- DNA Barcoding
实验技术流程:
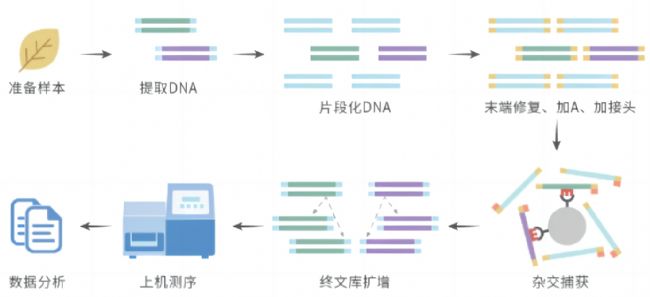
生信分析流程:
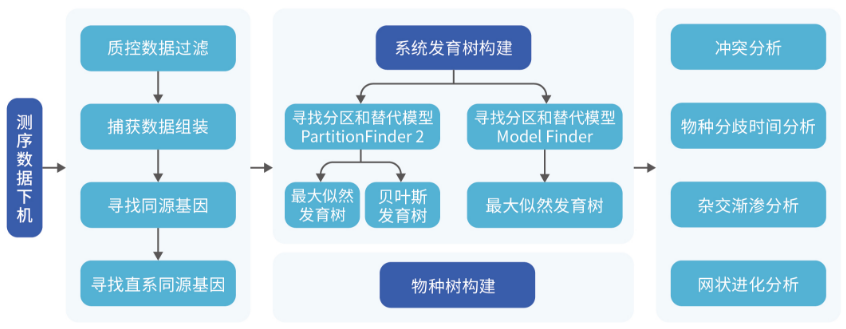
实验数据:
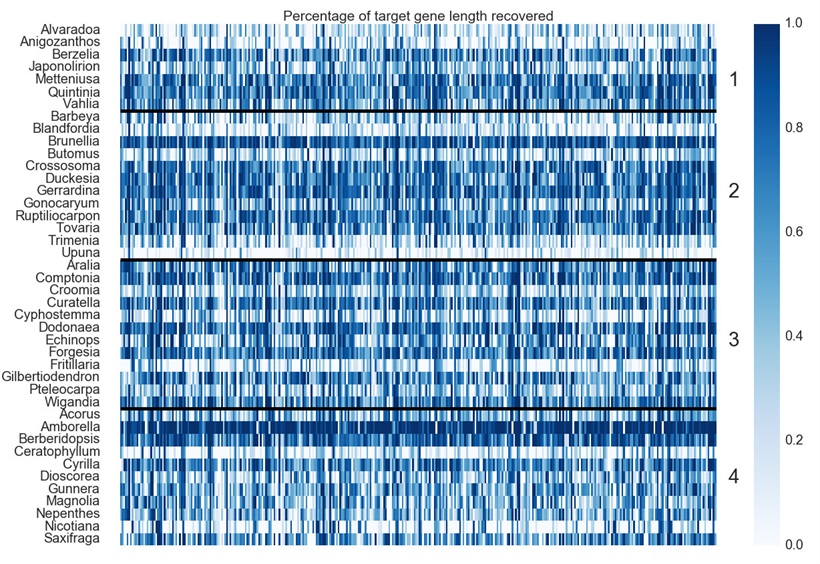
Fig 1. Heatmap of Gene Recovery Efficiency. Each row is one sample, and each column is one gene. Colors indicate the percentage of the target length (calculated by the mean length of all k-medoid transcripts for each gene) recovered.
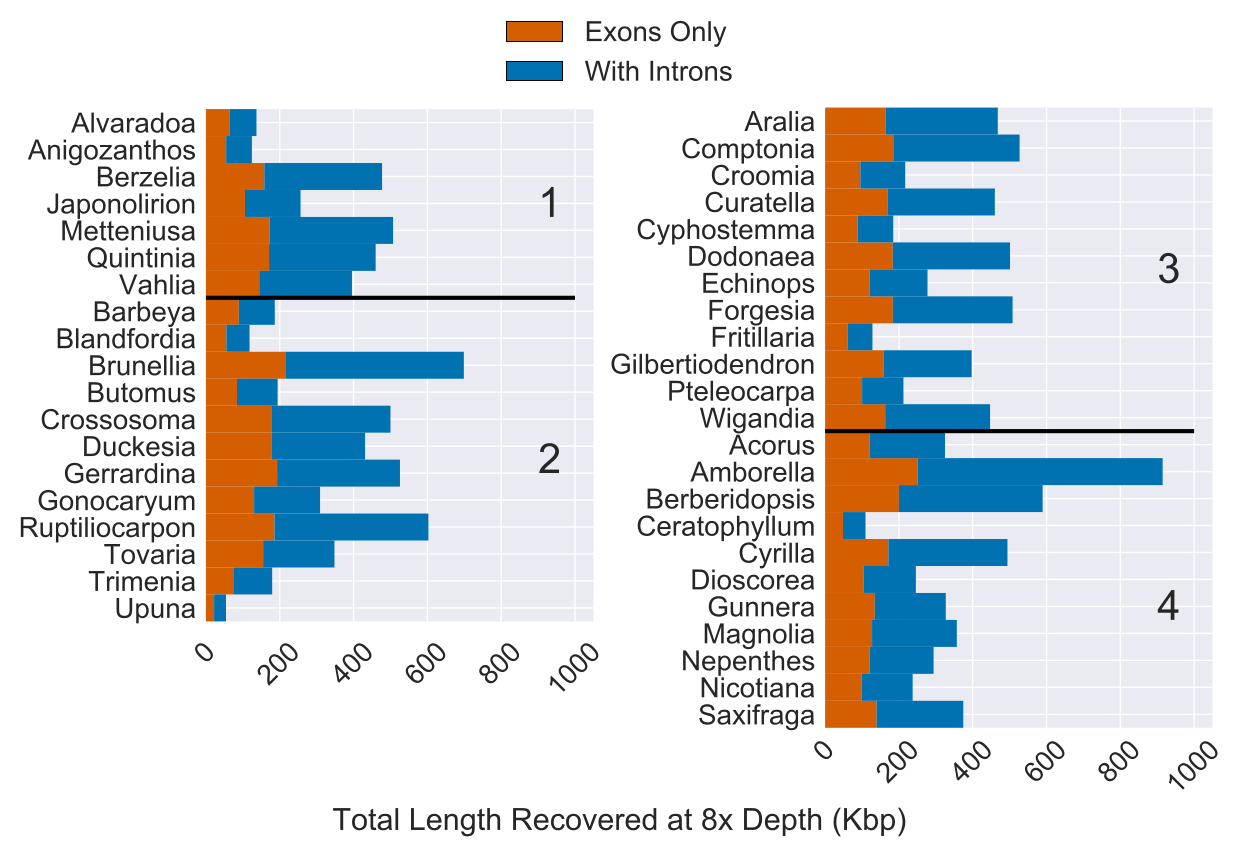
Fig 2. Total Length of Sequence Recovery for Both Coding and Non-coding Regions Across 353 Loci for 42 Angiosperm Species. Reads were mapped back to either coding sequence (yellow) or coding sequence plus flanking non-coding (i.e. intron) sequence (purple)… The total length of coding sequence targeted was 260,802 bp. The median recovery of coding sequence was 137,046 bp and the median amount of non-coding sequence recovered was 216,816 bp (with at least 8x depth of coverage).
参考文献:
Johnson MG, Pokorny L, Dodsworth S, Botigué LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT, Leebens-Mack JH, Leitch IJ, Maurin O, Soltis DE, Soltis PS, Wong GK, Baker WJ, Wickett NJ. A Universal Probe Set for Targeted Sequencing of 353 Nuclear Genes from Any Flowering Plant Designed Using k-Medoids Clustering. Syst Biol. 2019. 68(4):594-606. doi: 10.1093/sysbio/syy086.
蓝景科信为您提供被子植物353个单拷贝核基因靶向捕获测序全流程技术服务和个性化数据分析,及基于靶向捕获测序技术的基因组整体解决方案。欢迎咨询!






